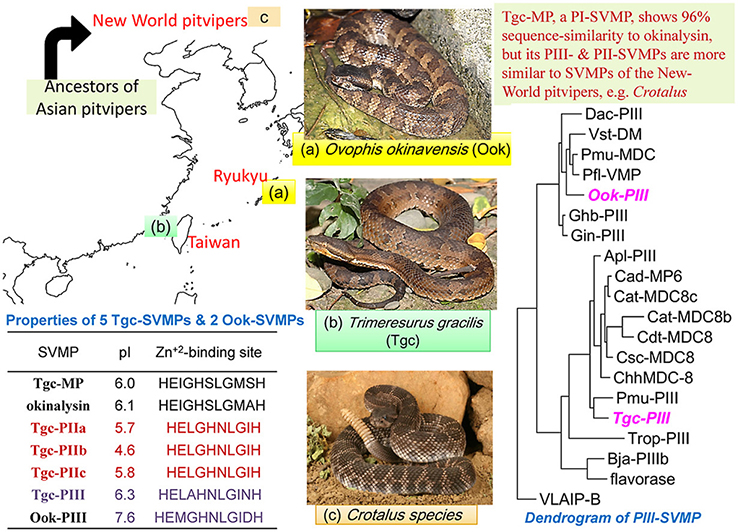
The cDNAs encoding the Zn+2-metalloproteases (SVMPs) of Trimeresurus gracilis (abbreviated as Tgc), a pitviper endemic to Taiwan, were cloned from venom glands and sequenced. The amino-acid sequences of five novel SVMPs, including one P-III, three P-II and one P-I class enzymes, were thus deduced and subjected to BLAST-analyses. The P-III enzyme (designated as Tgc-PIII) is structurally most similar to the PIII-SVMPs of New World pitvipers but not similar to the PIII-SVMP of Ovophis okinavensis. Sequence-similarity analysis of 22 homologous PIII-SVMPs reveal three major structural subtypes of the pitviper PIII-SVMPs, which possibly have different substrate specificities. In addition, Tgc-PIII and the PI-class SVMP (named Tgc-MP) were isolated from the venom and verified by mass spectrometry. All the three deduced sequences of PII-SVMPs (Tgc-PIIs) contain an abnormal Zn+2-binding-site in their catalytic-domain, and an identical "long-disintegrin" domain. The predicted 85-residues disintegrin, gracilisin, bears high similarities to some long-disintegrins of the New-World pitvipers and salmosin3. By BLAST search and comparison, Tgc-MP is 96% similar to okinalysin, the hemorrhagic PI-SVMP of O. okinavensis, rather than any other PI-SVMPs in the databanks. Our results confirm the fast evolution of Tgc-SVMPs as well as their structural similarities to different SVMP-classes of the New-World pitvipers and of O. okinavensis, respectively. The implications of our findings are discussed along with our previous sequence comparisons of venom phospholipases A2 and ten venom serine proteases of Tgc.

 Institute of Biological Chemistry, Academia Sinica
Institute of Biological Chemistry, Academia Sinica