中央研究院 生物化學研究所
中央研究院 生物化學研究所
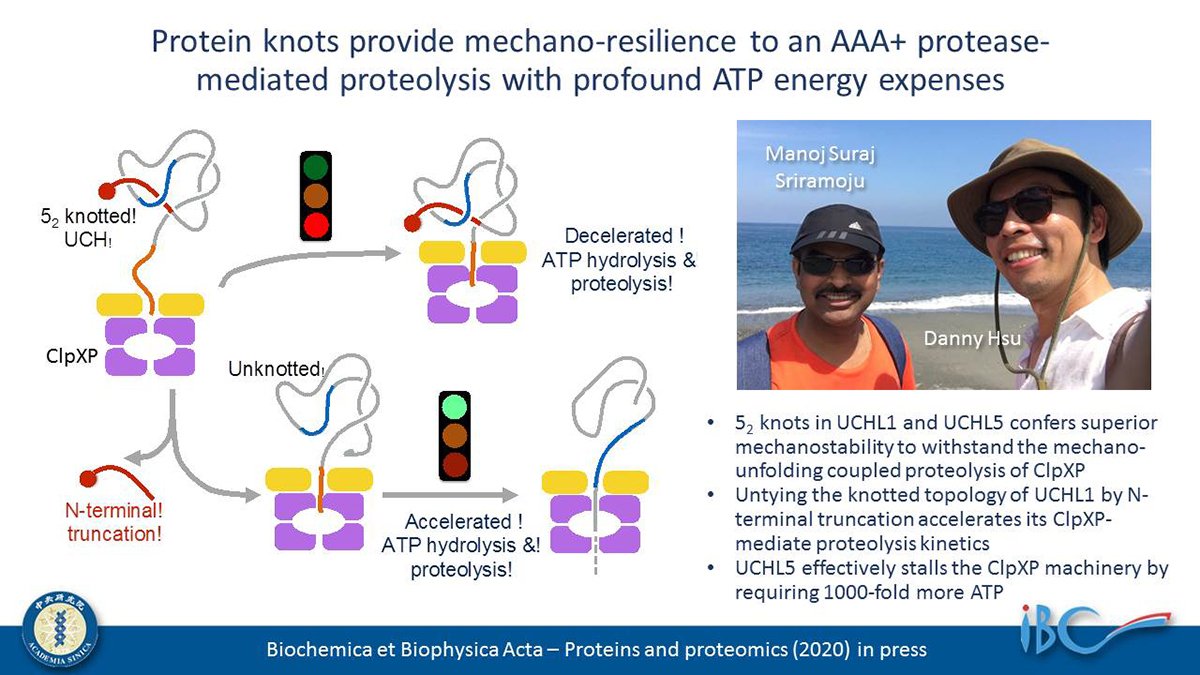
Knotted proteins are some of the most fascinating examples of how linear polypeptide chains can achieve intricate topological arrangements efficiently and spontaneously. The entanglements of polypeptide chains could potentially enhance their folding stabilities. We recently reported the unprecedented mechanostability of the Gordian (52) knotted family of human ubiquitin C-terminal hydrolases (UCHs) in the context of withstanding the mechanical unfolding of the bacterial AAA+ proteasome, ClpXP; a green fluorescence protein (GFP) was fused to the N-terminus of various UCHs as a reporter of the unfolding and degradation of these topologically knotted substrates, but it also limited the ability to examine the effect of untying the knotted topology via N-terminal truncation. In this study, we directly monitored the ClpXP-mediated degradation of UCH variants by electrophoresis and quantitative imaging analyses. We demonstrated that untying of the 52 knot in UCHL1 via N-terminal truncation (UCHL1Δ11) significantly reduces its mechanostability. We further quantified the ATP expenditures of degrading different UCH variants by ClpXP. The unknotted UCHL1Δ11 underwent accelerated ClpXP-dependent proteolysis, with a 30-fold reduction in ATP consumption compared to the knotted wild type. Unlike all other known ClpXP substrates, UCHL5, which is the most resilient substrate known to date, significantly slowed down the ATP turnover rate by ClpXP. Furthermore, UCHL5 required 1000-fold more ATP to be fully degraded by ClpXP compared to GFP. Our results underscored how the complex, knotted folding topology in UCHs may interfere with the mechano-unfolding processes of the AAA+ unfoldase, ClpX.