中央研究院 生物化學研究所
中央研究院 生物化學研究所
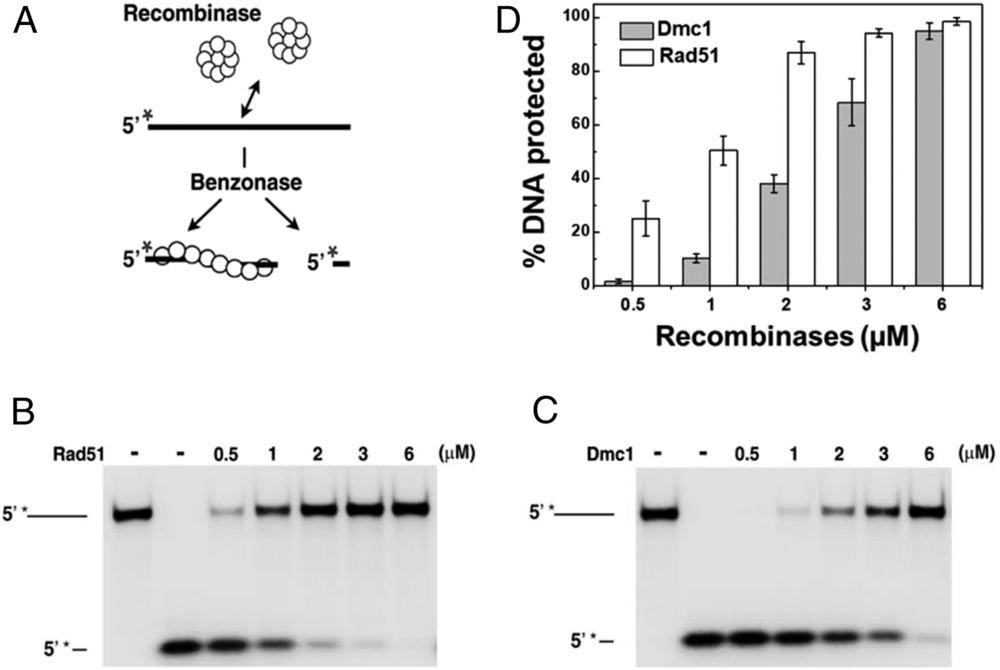
Dmc1 recombinases are essential to homologous recombination in meiosis. Here, we studied the kinetics of the nucleoprotein filament assembly of Saccharomyces cerevisiae Dmc1 using single-molecule tethered particle motion experiments and in vitro biochemical assay. ScDmc1 nucleoprotein filaments are less stable than the ScRad51 ones because of the kinetically much reduced nucleation step. The lower nucleation rate of ScDmc1 results from its lower single-stranded DNA (ssDNA) affinity, compared to that of ScRad51. Surprisingly, ScDmc1 nucleates mostly on the DNA structure containing the single-stranded and duplex DNA junction with the allowed extension in the 5'-to-3' polarity, while ScRad51 nucleation depends strongly on ssDNA lengths. This nucleation preference is also conserved for mammalian RAD51 and DMC1. In addition, ScDmc1 nucleation can be stimulated by short ScRad51 patches, but not by EcRecA ones. Pull-down experiments also confirm the physical interactions of ScDmc1 with ScRad51 in solution, but not with EcRecA. Our results are consistent with a model that Dmc1 nucleation can be facilitated by a structural component (such as DNA junction and protein-protein interaction) and DNA polarity. They provide direct evidence of how Rad51 is required for meiotic recombination and highlight a regulation strategy in Dmc1 nucleoprotein filament assembly.