中央研究院 生物化學研究所
中央研究院 生物化學研究所
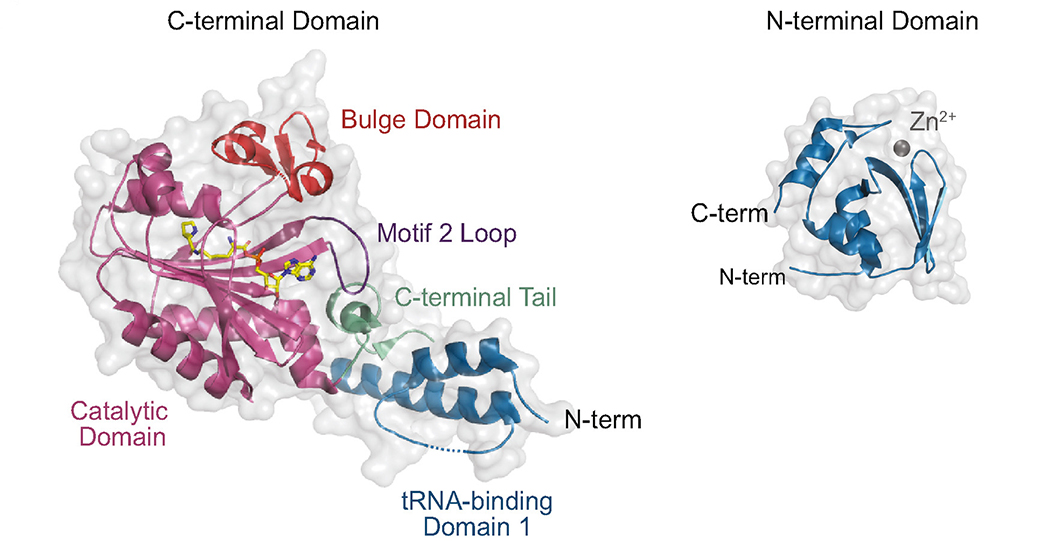
The pyrrolysyl-tRNA synthetase (PylRS) facilitates the co-translational installation of the 22nd amino acid pyrrolysine. Owing to its tolerance for diverse amino acid substrates, and its orthogonality in multiple organisms, PylRS has emerged as a major route to install noncanonical amino acids into proteins in living cells. Recently, a novel class of PylRS enzymes was identified in a subset of methanogenic archaea. Enzymes within this class (ΔPylSn) lack the N-terminal tRNA-binding domain that is widely conserved amongst PylRS enzymes, yet remain highly active and orthogonal in bacteria and eukaryotes. In this study, we use biochemical and in vivo UAG-readthrough assays to characterize the aminoacylation efficiency and substrate spectrum of a ΔPylSn class PylRS from the archaeon Ca. Methanomethylophilus alvus. We show that, compared to the full-length enzyme from Methanosarcina mazei, the Ca. M. alvus PylRS displays reduced aminoacylation efficiency, but an expanded amino acid substrate spectrum. To gain insight into the evolution of ΔPylSn enzymes, we performed molecular phylogeny using 156 PylRS and 105 tRNAPyl sequences from diverse anaerobic archaea and bacteria. This analysis suggests that the PylRS•tRNAPyl pair diverged before the evolution of the three domains of life, placing an early limit on the evolution of the Pyl-decoding trait. Furthermore, our results document the co-evolutionary history of PylRS and tRNAPyl and reveal the emergence of tRNAPyl sequences with unique A73 and U73 discriminator bases. The orthogonality of these tRNAPyl species with the more common G73-containing tRNAPyl will enable future efforts to engineer PylRS systems for further genetic code expansion.