中央研究院 生物化學研究所
中央研究院 生物化學研究所
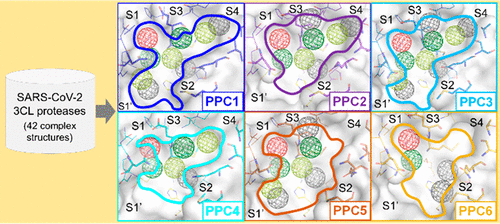
The infectious SARS-CoV-2 causes COVID-19, which is now a global pandemic. Aiming for effective treatments, we focused on the key drug target, the viral 3C-like (3CL) protease. We modeled a big dataset with 42 SARS-CoV-2 3CL protease-ligand complex structures from ∼98.7% similar SARS-CoV 3CL protease with abundant complex structures. The diverse flexible active site conformations identified in the dataset were clustered into six protease pharmacophore clusters (PPCs). For the PPCs with distinct flexible protease active sites and diverse interaction environments, we identified pharmacophore anchor hotspots. A total of 11 "PPC consensus anchors" (a distinct set observed in each PPC) were observed, of which three "PPC core anchors" EHV2, HV1, and V3 are strongly conserved across PPCs. The six PPC cavities were then applied in virtual screening of 2122 FDA drugs for repurposing, using core anchor-derived "PPC scoring S" to yield seven drug candidates. Experimental testing by SARS-CoV-2 3CL protease inhibition assay and antiviral cytopathic effect assays discovered active hits, Boceprevir and Telaprevir (HCV drugs) and Nelfinavir (HIV drug). Specifically, Boceprevir showed strong protease inhibition with micromolar IC50 of 1.42 μM and an antiviral activity with EC50 of 49.89 μM, whereas Telaprevir showed moderate protease inhibition only with an IC50 of 11.47 μM. Nelfinavir solely showed antiviral activity with a micromolar EC50 value of 3.28 μM. Analysis of binding mechanisms of protease inhibitors revealed the role of PPC core anchors. Our PPCs revealed the flexible protease active site conformations, which successfully enabled drug repurposing.