中央研究院 生物化學研究所
中央研究院 生物化學研究所
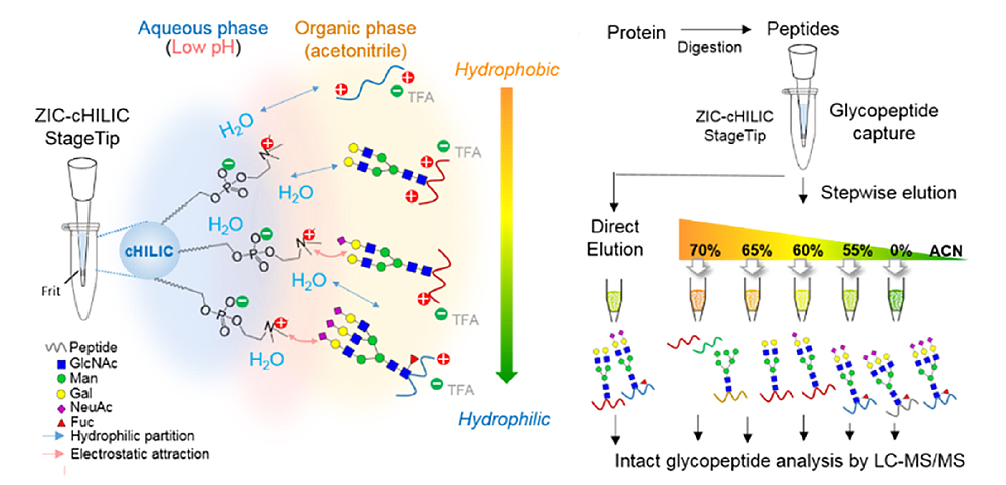
Alterations of protein glycosylation are closely related with pathophysiological regulation. Due to the structural macro- and microheterogeneity, low stoichiometry, and low ionization efficiency of glycopeptides, high-performance tools to enrich glycopeptides, especially the negatively charged and labile sialoglycopeptides, are essential to enhance the identification of the underexplored glycoproteome. Here, we present the first implementation of zwitterionic hydrophilic interaction chromatography with the exposed choline group (ZIC-cHILIC) in StageTip for simultaneous enrichment and fractionation of intact glycopeptides. In a model study using lung cancer cells, early elution by a high percentage of acetonitrile prominently prefilters nonglycopeptides, facilitating high enrichment specificity for glycopeptides (92-96%) and sialoglycopeptides (77-89%) in the subsequent hydrophilic fractions. The stepwise elution shows a high glycopeptide fractionation efficiency by a <10% overlap of glycopeptides between adjacent fractions. Most importantly, the ZIC-cHILIC stepwise strategy demonstrated good reproducibility (>80% in triplicate analysis) as well as superior coverage of 4.6- to 12.0-fold and 2.1- to 35.6-fold more glycopeptides and sialoglycopeptides compared to conventional TiO2 and ZIC-HILIC, respectively. To the best of our knowledge, the result with 2742 sialoglycopeptides among 7367 unique glycopeptides and 166 glycans from 2434 N-glycosites of 1118 glycoproteins (Byonic score > 100) provides one of the deepest glycoproteomic profiles in single-cell type. Without the immunoprecipitation step, the large-scale glycoproteomic atlas also reveals site-specific glycosylation of many druggable receptor proteins, such as EGFR, MET, ERBB2, ERBB3, AXL, and IGF1R. The demonstrated high enrichment specificity and identification depth show that stepwise ZIC-cHILIC is an efficient method to explore the under-represented sialoglycoproteome.