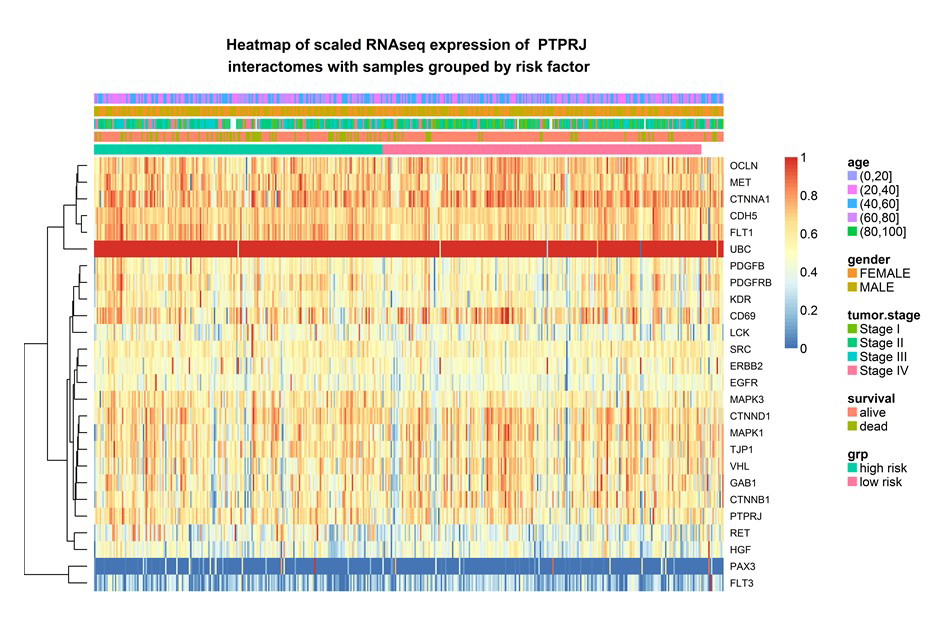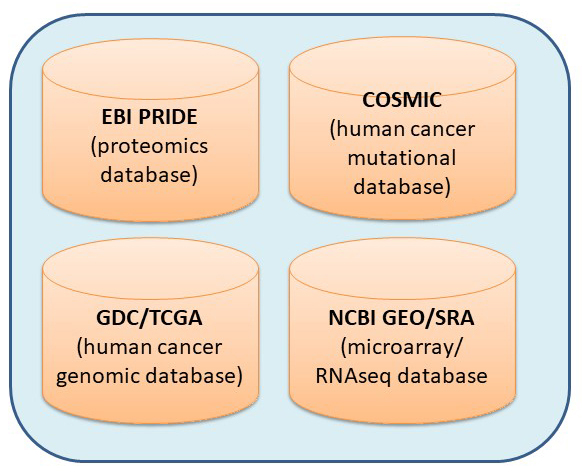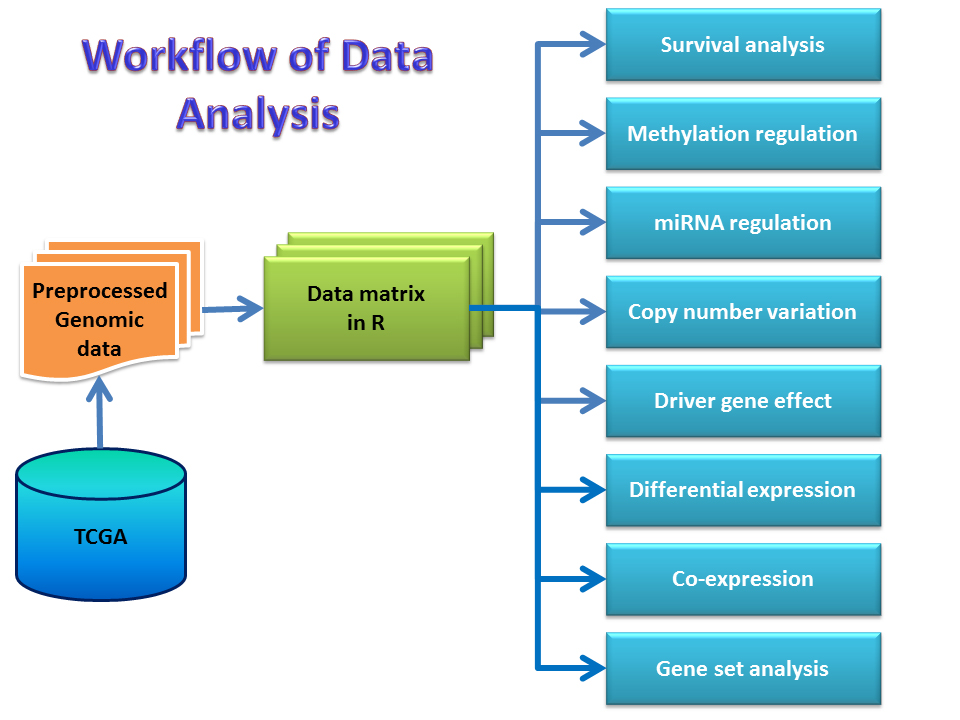
The core installs various servers to meet different needs of bioinformatics.The in-house galaxy server is a web-based platform for small-scale data analysis with user-friendly interface and wide-range of bioinformatics tools. The Linux server has the specifications for high-throughput data analysis from next generation sequencing and genome assembly. Major tools installed on the server include SPAdes/MaSuRCA for genome assembly, maker for genome annotation, Salmon for RNAseq quantation, Blast+ on NCBI nr database, AntiSMASH for fungi/bacteria for gene cluster analysis, Mothur for 16S rRNA sequence data analysis, etc. The web server allows building in-house web applications and R/Shiny server supports the R-based statistics and interactive graphs for in-house web applications.